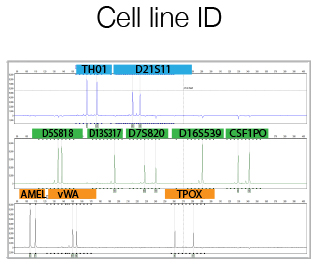
Overview
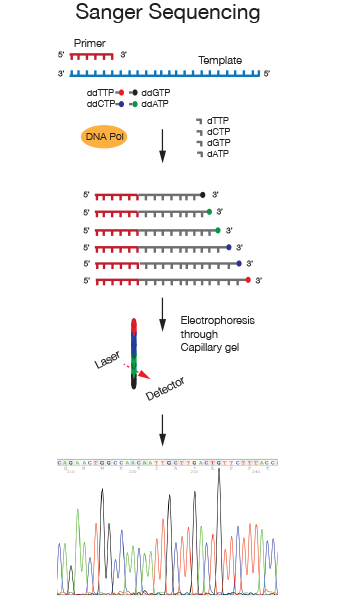
The DNA Sanger Sequencing Service was established in the early 2000s, with the introduction of automated capillary instrumentation and BygDye fluorescent terminator chemistry to ensure high quality reads from different types of templates.
In more than 20 years of experience in this field, we have provided technical and scientific support to numerous basic and translational research projects.
In addition to this, the DNA Sanger Sequencing Service collaborates at the forefront of the diagnostic activities of the
CGT lab.
Specifically, for DNA sequencing we offer:
- storage of samples for up to 1 month
- free universal primers
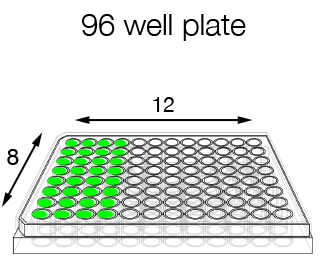
- 96-well format (for homogeneous samples and to be processed with the same oligo) and single-tube format
- Ready-to-go format, in which the customer provide pre-mixed DNA and primers
- assistance
- delivery of results in 24-48 hours
- free reloading
- shipment of data in .ab1 format
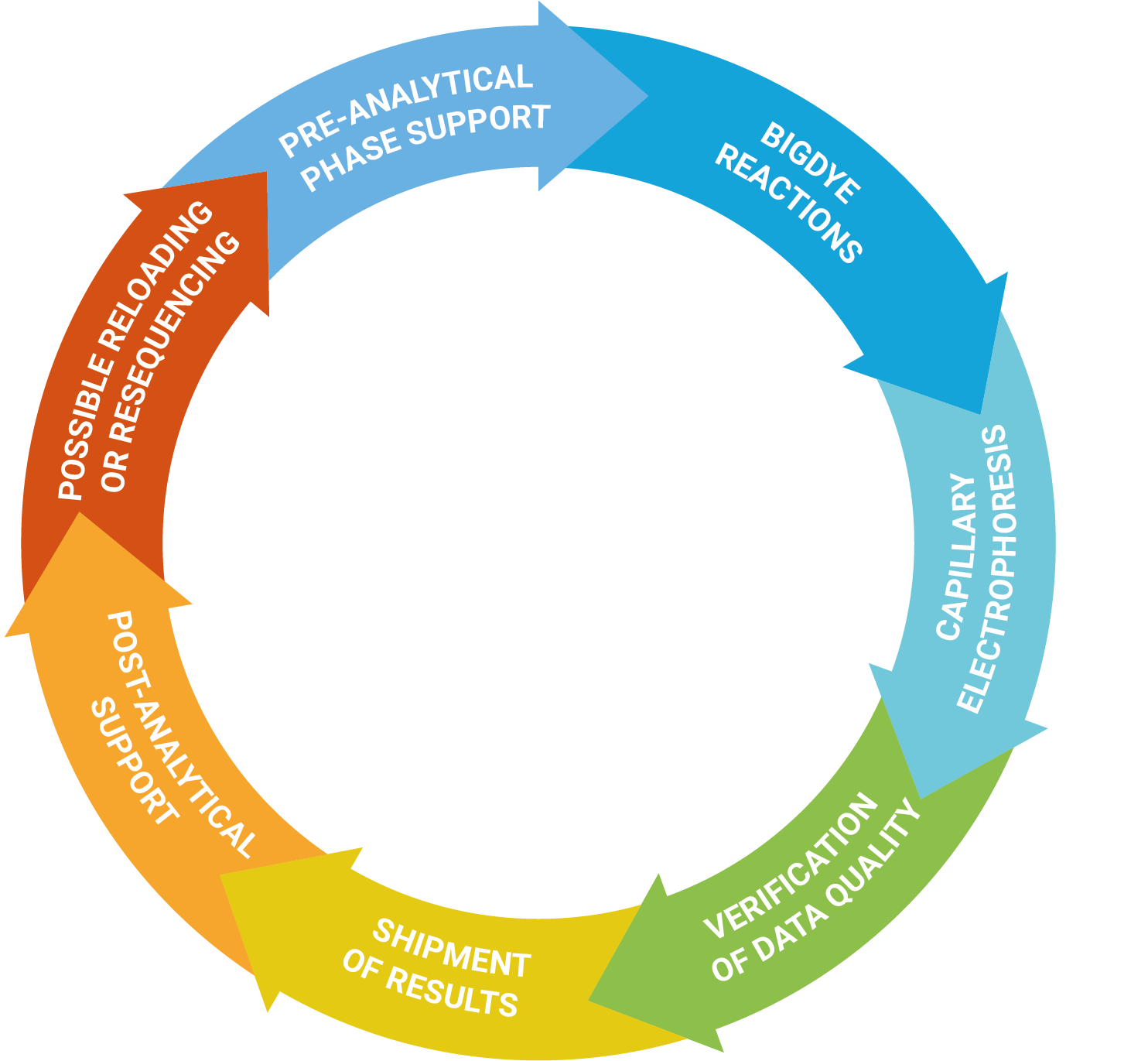
Service
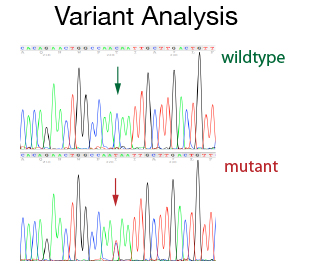
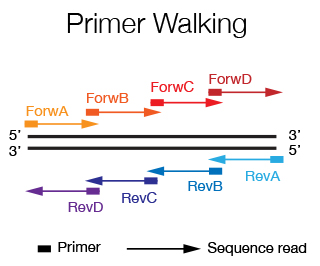
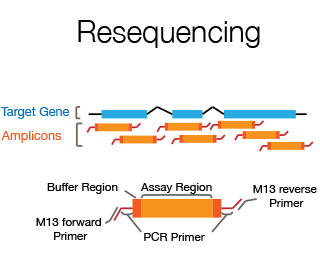
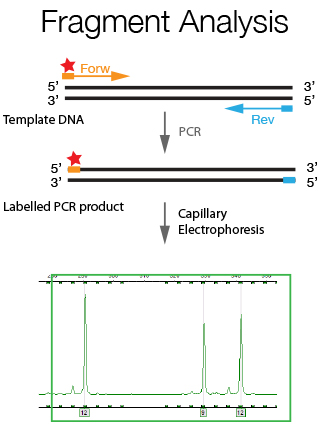
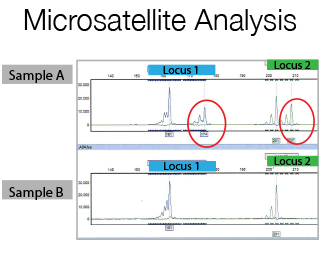
The DNA Sanger Sequencing unit can offer standard DNA sequencing and assistance for projects using on Sanger sequencing. We also offer the possibility to use fragment analysis for various applications, providing assistance in the pre-and post-analytical phase.
Innovation
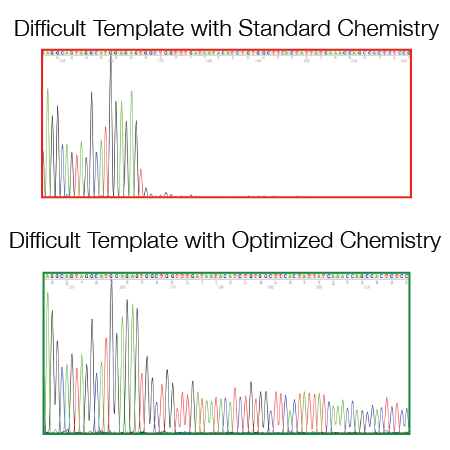
As a routine practice, we revise existing protocols related to the Sanger sequencing method, following updates in technology and reagents, to optimize and improve our work and the offer provided to our clients.
We maintain a constantly updated and optimized list of free universal primers.
Quality
The laboratory is ISO 9001 accredited.
We provide a sequencing read capacity of 600-800 base pairs, with 99.99% accuracy (Q 40).
UNI EN ISO 9001:2015 n. IT324391

Services
FAQ e Technical Info
Staff and Contacts

Sara Volorio
Sara Volorio graduated in Biology in 1995 from the University of Milan. She worked for 5 years at the TIGEM Institute where she discovered her passion for sequencing. She then moved in the year 2000 to IFOM where she later became the head of the DNA Sanger Sequencing Unit. She is also involved in R&D for the CGT Lab.

Stefano Fortuzzi
Stefano Fortuzzi graduated in 2005 in Agricultural and Plant Biotechnology from the University of Bologna. Subsequently he started working at Cogentech in the Genetic Testing group where he still develops and implements the automation of workflows. In 2014 he joined the DNA Sanger Sequencing Unit where he still works today.

Domenico Sardella
Domenico Sardella in 2003 received a University Diploma in Biomedical Laboratory Health Technician from the University of Urbino. He worked at the National Cancer Institute and the European Institute of Oncology, before joining the Cogentech DNA sanger Sequencing Service in 2012.
Contacts
- Phone
+39 02 574303210
+39 02 574303207 (lab) - Email
sequencing-service [@] cogentech.it